FileNotFoundError: [Errno 2] No such file or directory: '../data/2011_Kling_Akchem.csv'Return to Course Home Page
TryPy 2 - For Loops and Conditionals
Part 0. Setup Steps
- Create a repo on GitHub named
eds217-trypy-02 - Clone to create a version-controlled project
- Create some subfolder infrastructure (nbs, data, etc..)
- Create a new python notebook.
Part 1. Real data
Explore this data package from EDI, which contains a “Data file describing the biogeochemistry of samples collected at various sites near Toolik Lake, North Slope of Alaska”. Familiarize yourself with the metadata (particularly, View full metadata > expand ‘Data entities’ to learn more about the variables in the dataset).
Citation: Kling, G. 2016. Biogeochemistry data set for soil waters, streams, and lakes near Toolik on the North Slope of Alaska, 2011. ver 5. Environmental Data Initiative. https://doi.org/10.6073/pasta/362c8eeac5cad9a45288cf1b0d617ba7
- Download the CSV containing the Toolik biogeochemistry data
- Take a look at it - how are missing values stored? Keep that in mind.
- Drop the CSV into your data folder of your project
- Create a new notebook document in VSCode (or in a
jupyter notebookserver), save in docs astoolik_chem.ipynb - Import the
pandas,numpy, andmatplotlib.pyplotlibraries into your first code cell. - Read in the data as
toolik_biochem. Remember, you’ll want to specify here howNAvalues are stored (hint: use thena_valuesargument in yourpd.read_csvcall.) Additionally, convert all column names to lower case/underscore format (replace spaces with underscores and put everything in lower case).
Create a subset of the data that contains only observations from the “Toolik Inlet” site, and that only contains the variables (columns) for pH, dissolved organic carbon (DOC), and total dissolved nitrogen (TDN). Store this subset as
inlet_biochem. Make sure to LOOK AT the subset you’ve created.Find the mean value of each column in
inlet_biochem2 different ways:
- Write a for loop from scratch to calculate the mean for each
- Use one other method (e.g.
.mean(), or.apply()) to find the mean for each column.
Using for loop:
col ph: 7.06
col doc_um: 409.96
col tdn_um: 13.37
Using list comprehension:
col ph: 7.06
col doc_um: 409.96
col tdn_um: 13.37
Using df.mean()
ph 7.063182
doc_um 409.961538
tdn_um 13.365385
dtype: float64
Using .apply()
ph 7.063182
doc_um 409.961538
tdn_um 13.365385
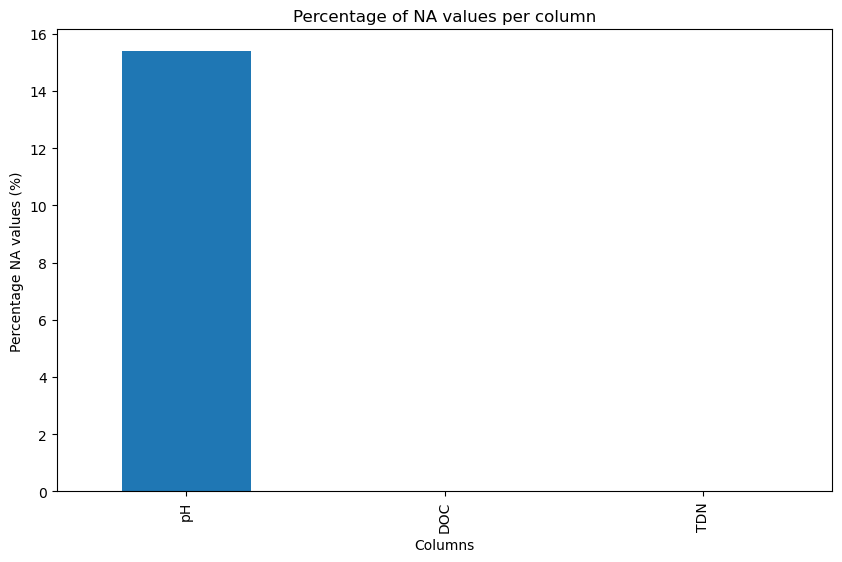
dtype: float64- Count the number of na values you have for each column and create a bar chart showing the % na’s for each.
Code
# Count the number of NA values for each column
na_counts = inlet_biochem.isna().sum()
# Calculate the percentage of NA values for each column
total_rows = len(inlet_biochem)
na_percentage = (na_counts / total_rows) * 100
# Plotting
plt.figure(figsize=(10, 6))
ax = na_percentage.plot(kind='bar')
plt.title('Percentage of NA values per column')
plt.xlabel('Columns')
plt.ylabel('Percentage NA values (%)')
# Rename x-axis labels
new_labels = ['pH', "DOC", "TDN"]
ax.set_xticklabels(new_labels)
plt.show()